I want to introduce RJMCMC and demonstrate how it can be programmed in Stata. To understand this algorithm, you really do need to know a little about the basis of Metropolis-Hastings, so this week I will run through the ideas behind the equations. A bit of algebra I am afraid, but once it is out of the way, we can return to practical applications.
A few weeks ago I looked at the Google Flu Trends data and fitted a Bayesian model that identifies the start and end of the annual flu epidemic. A limitation of that model is that it must have exactly one epidemic every year. This is unrealistic as there is a clear suggestion that in some years we do not see a flu epidemic at all and in other years there appear to be two distinct outbreaks.
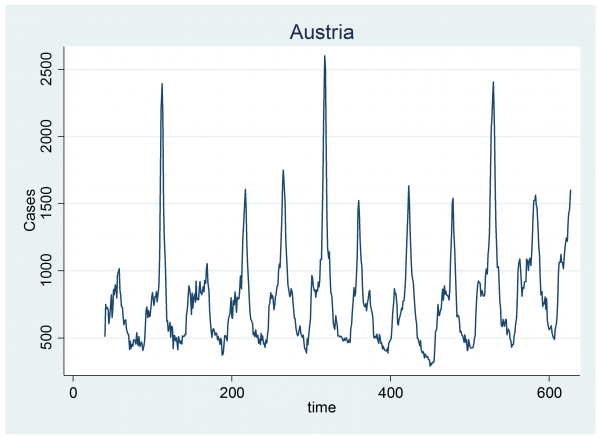
These patterns can be seen in the data for Austria.
When we allowed exactly one epidemic per year, the Poisson model for the number of cases per week had an expected number, mu, where,
log(mu) = a + b time/1000 + c sin(0.060415*week)+Σ Hiδ(year==i & week>=Si & week<=Ei)
and (Hi,Si,Ei) represent the height, start and end of the single epidemic in year i and we sum over the different years.
What I want to do now is to develop a model that it is able to allow the number of epidemics per year to vary. Suppose that we were to generalize so that there could be either 0, 1 or 2 epidemics in any year. We have 12 years of data from Austria, so there will be 312=531,441 possible models each with a unique combination of 0, 1 or 2 epidemics in the different years.
Since we cannot be certain which of the 531,441 models will apply, we need to develop a Bayesian analysis that will select the most appropriate model as part of the fitting process and this means that our MCMC chain must move freely between the models. The main obstacle to developing such an algorithm is that, as we move from one model to another, so the number of parameters will change with the number of epidemics. We will need to develop a version of the Metropolis-Hastings algorithm that can cope with this problem.
The most commonly used algorithm for this type of Bayesian model selection is called reversible jump Markov chain Monte Carlo or RJMCMC.
This week I want to outline the theory behind RJMCMC and then next week I’ll take a simple example and program it in Stata. Eventually, I want to return to the Austrian flu data and see if I can produce a Stata program to fit a Bayesian model to those data. I must admit that I have not tried this yet and I suspect that selection from half-a-million possible models might prove quite a challenge.
When we allow up to 2 epidemics per year, the Poisson model will have three parameters that control the pattern outside of an epidemic, namely, the intercept, the coefficient of the linear time trend and the coefficient of the sine term. On top of these, there will be a further three parameters for each epidemic (start, end and height). So, in the extreme case, if there were 2 epidemics in every one of the 12 years, we would require 3+12*2*3=75 parameters. Let’s imagine a vector Ψ that contains all 75.
For a particular model we might have one epidemic during most years but occasionally no epidemic or two epidemics, so often we will need some of the 75 parameters and not others. Let’s suppose that for model Mk we will need parameters θk and we will not need parameters uk, where θk and uk together make up Ψ.
Now imagine that the chain is currently in Model k and we are considering a move to Model j, i.e. from parameters (θk,uk) to (θj,uj).
Our aim is to generate a chain in which we visit model Mk with the posterior probability P(Mk|Y,Ψ), where Ψ is the current value of the parameters. Averaging over the whole chain will be like integrating over Ψ and will mean that the chain is in model k with probability P(Mk|Y).
If the chain really does have model probabilities P(Mk|Y,Ψ) and it is constructed by movements between models that have transition probabilities such as P(move Mj to Mk) then for the chain to stay converged we will require that,
Sumj P(Mj|Y,Ψ) P(move Mj to Mk) = P(Mk|Y,Ψ)
Which, in words, means that the total chance of moving into Model k must be equal to the probability of model k.
One way to ensure that this property holds is to have detailed balance, which says that, the probability of being in Model k and moving to Model j must equal the chance of being in Model j and moving to Model k.
P( Mk|Y, Ψ) P(move Mk to Mj) = P( Mj|Y, Ψ) P(move Mj to Mk)
If detailed balance holds then,
Sumj P(Mj|Y,Ψ) P(move Mj to Mk) = Sumj P( Mk|Y, Ψ) P(move Mk to Mj)
and as the sum of the probabilities of all of the allowed moves from Model k must be one,
Sumj P(Mj|Y,Ψ) P(move Mj to Mk) = P( Mk|Y, Ψ) Sumj P(move Mk to Mj) = P(Mk|Y, Ψ)
which is just what we need.
In Metropolis-Hastings algorithms, movement between models is a two stage process made out of a proposal and its acceptance or rejection. So detailed balance becomes,
P( Mk|Y, Ψ) g(Mk to Mj) a(Mk to Mj) = P( Mj|Y, Ψ) g(Mj to Mk) a(Mj to Mk)
where I have used g to represent the proposal probability and a to represent the acceptance probability.
This tells us that, at any point in the chain, the ratio of the acceptance probabilities a(Mk to Mj)/ a(Mj to Mk) must equal
P(Mj|Y, Ψ) g(Mj to Mk) / P(Mk|Y, Ψ) g(Mk to Mj)
In essence this is the Metropolis-Hastings algorithm for movement between Models. We calculate this ratio based on the posterior P(Mk|Y, Ψ) and our personal choice of movement probabilities g(). Then any pair of acceptance probabilities that have this ratio will create a chain that converges to the correct posterior, so such a chain will select the different models with frequencies that are proportional to the posterior model probabilities.
Since we want to encourage moves in order to obtain good mixing, it makes sense to make the larger of the two acceptance probabilities equal to one. For instance, if the larger is a(Mj to Mk) then we set it to 1 and make,
a(Mk to Mj) = P(Mj|Y, Ψ) g(Mj to Mk) / P(Mk|Y, Ψ) g(Mk to Mj)
and if a(Mk to Mj) is the larger then we set it to 1 and make,
a(Mj to Mk) = P(Mk|Y, Ψ) g(Mk to Mj) / P(Mj|Y, Ψ) g(Mj to Mk)
This can be summarised as
a(Mj to Mk) = min { P( Mk|Y, Ψ) g(Mk to Mj) / P( Mj|Y, Ψ) g(Mj to Mk) , 1}
which is how people usually write the M-H acceptance probability.
All we need to do now is to decide how to calculate P(Mk|Y, Ψ). Bayes theorem tells us that
P(Mk|Y,Ψ) α P(Y|Ψ,Mk) P(Mk|Ψ) α P(Y|Ψ,Mk) P(Ψ|Mk) P(Mk)
So
a(Mj to Mk) = min { P(Y|Ψ,Mk) P(Ψ|Mk) P(Mk) g(Mk to Mj) / P(Y|Ψ,Mj) P(Ψ|Mj) P(Mj) g(Mj to Mk) , 1}
This looks like a standard MH algorithm. So where is the problem? In fact there three hidden issues.
(a) we do not use Ψ in our model specification but a subset of Ψ. Remember that Ψ=(Θ,u) and the split will not be the same for Mk and Mj.
(b) the algebraic forms of the models Mk and Mj may be different (e.g. a gamma and a log-normal), in which case the constants in the formulae may not cancel as they do in most MH calculations.
(c) Bayes theorem applies to probabilities, P, but our models are usually expressed as probability densities, f. Since the area under f defines probability, you might say that P(Ψ|Mk) = f(Ψ|Mk) dΨ where dΨ defines a tiny region with the same dimension as Ψ. So the acceptance probabilities ought to be written as
a(Mj to Mk) = min { f(Y|Ψ,Mk)dY f(Ψ|Mk) dΨ P(Mk) g(Mk to Mj) / f(Y|Ψ,Mj)dY f(Ψ|Mj) dΨ P(Mj) g(Mj to Mk) , 1}
Notice that there is no small increment associated with Mk because the models are discrete and are described directly by probabilities and the delta for Y will cancel as the data are the same whichever model we use.
The reason that we do not usually bother with the dΨ is that, in many circumstances, it too cancels. Unfortunately we have to be very careful over this cancellation when the parameterizations of the two model are different. In that case, the ratio of the deltas becomes the Jacobian of the transformation from one set of parameters to the other.
We will sort out the specifics of this calculation when we consider particular examples.
One question that we have not yet addressed is how we choose the moves; a choice that is equivalent to defining the function g(). As with any MH algorithm this choice is arbitrary provided that a possible move from Mk to Mj is matched by a possible move from Mj to Mk and we do not allow cul-de-sacs from where it is impossible to reach some of the other models. So the algorithm will work for a more or less arbitrary g() but some choices will lead to very poorly mixing chains in which we stay with the same model for a long time before we switch, while other types of proposals will lead to frequent moves and better mixing. Part of the skill of using RJMCMC is to come up with an efficient strategy for proposing moves between models.
In the case of the epidemic models, we could have a scheme that proposes a move from Mk to a random choice among the 533,540 possible alternative models but such an algorithm would be incredibly difficult to program and would lead to a lot of rejections, so it would be hugely inefficient. An algorithm in which we make local moves is likely to perform much better. Perhaps, at any stage in the chain, we could select a year at random and either propose that we add one epidemic or drop one epidemic. Once again this is best illustrated in the context of a specific example.
So next week I will take an example, fill in the details and then program the algorithm in Stata.
I’ll finish this week with a reference. The original idea of RJMCMC was developed by Peter Green in 1995 (Biometrika, 82(4), 711-732). That paper is surprisingly readable for Biometrika but it is probably not the best introduction to the topic. My suggestion is to start with the simplified approach described in,
Barker, R. J., & Link, W. A. (2013).
Bayesian multimodel inference by RJMCMC: a Gibbs sampling approach.
The American Statistician, 67(3), 150-156.


 Subscribe to John's posts
Subscribe to John's posts
Recent Comments