This week I am going to present a program that runs Stan from within Stata, but in this version, there will be a minimum of automation. Next time, I will modify the wbs programs, designed for running WinBUGS, so that they will also work with Stan and at that stage the process will be simpler to use but less transparent.
We need a simple problem, so let us stick with the Bernoulli data problem introduced last week, but we will simulate our own data so that we can vary the number of observations and the true value of theta.
. clear
. set obs 20
. local TRUE_THETA 0.4
. gen y = runiform() < `TRUE_THETA’
We need to write a model file using the Stan syntax. The manual describing this syntax is hundreds of pages long but because of the similarity between this problem and the Stan example all we need to do is to modify the model file provided by Stan.
The key difference that you will find in moving from WinBUGS to Stan is that Stan requires you to define the data and the parameters before you give the model. So our model file will have three blocks; data, parameters and model.
As well as providing the Stan model file, we also need to export the data, write a batch file of commands for running Stan and inspect the results. Since there is little automation, it should be quite easy for you to see what is involved in each of the stages. The full Stata do file is,
*--------------------------------------- * Simulate the data with theta=0.4 *--------------------------------------- clear set obs 20 local TRUE_THETA 0.4 gen y = runiform() < `TRUE_THETA' *--------------------------------------- * Write the model file: no problem *--------------------------------------- wbsmodel thisdofile.do BernModel.stan /* data { int N; int y[N]; } parameters { real theta; } model { theta ~ beta(1,1); for( i in 1:N ) { y[i] ~ bernoulli(theta); } } */ *--------------------------------------- * Write the data file: just like jags *---------------------------------------
wbslist_v2 (N <- 20) (vect y , format(%3.0f)) using BernData.txt, replace jags type BernData.txt *--------------------------------------- * Prepare a Windows batch file * wbsscript cannot do it as yet so write it directly * we will rely on Stan's defaults *--------------------------------------- wbsmodel thisdofile.do BernScript.bat /* c: cd c:/software/cmdstan-2.6.2 make k:/blog/topics/stan/BernModel.exe k: cd k:/blog/topics/stan BernModel.exe sample data file=BernData.txt */ *--------------------------------------- * Run the batch file *--------------------------------------- !BernScript.bat > log.txt type log.txt
*--------------------------------------- * By default the output is in output.csv * because it has a complex structure insheet etc will not read it *--------------------------------------- type output.csv
We still have a long way to go, but this program does work.
Let’s go through it, so as to understand what it does and to identify places where more work is needed in order to fully automate the running of Stan.
The model file
The model is written to a file called BernModel.stan using wbsmodel, just as we do with WinBUGS. We have to use a .stan extension for the file name so that the file will be recognised by Stan.
The contents of the file are more or less self-explanatory. The model block is similar to the model file for WinBUGS, although there are small differences such as beta() for the distribution rather than dbeta(). The main difference lies in the need for data and parameter blocks; WinBUGS deduces this information from the contents of the data file, while Stan requires it to be given explicitly (remember that Stan compiles the model before it sees the data). I’ll dedicate a full posting to the syntax once we have the procedure for working through Stata completed.
The data file
The format of the data file is identical to that used by JAGS except that the JAGS examples place the variable names in double quotes. The use of quotes is optional in JAGS so I have changes wbslist_v2 so that variable names are not quoted when the JAGS option is specified. Now we can create Stan data files by specifying the jags option. Eventually I will add a stan option to wbslist_v2 although it will not do anything different from the jags option.
The script file
We are working towards the automatic creation of the script but at the moment we have to write it out in full and copy it to a text file using wbsmodel. The file extension .bat identifies the file as a batch file of DOS commands. As it happens, I prepare the programs for this blog on an external drive so that I can test them on different computers. The cd command in the Command Prompt will only move between folders on the same drive, so we need commands like c: and k: to switch drives. Otherwise the commands are the same as we typed into the command prompt in last week’s posting.
Running the script
Eventually running the script will be a job for wbsrun, but at present the script is sent to the operating system using Stata’s ! command. The output from the job that would by default appear in the command prompt window, is diverted into a file called log.txt. When the job runs, a command prompt window opens and you will see a delay while the program is compiled and linked, but once it is ready to run, the results are obtained almost instantaneously.
The log file
Here is a listing of the log file (I’ve edited out some of the content where it is repetitive)
method = sample (Default)
sample
num_samples = 1000 (Default)
num_warmup = 1000 (Default)
save_warmup = 0 (Default)
thin = 1 (Default)
adapt
engaged = 1 (Default)
gamma = 0.050000000000000003 (Default)
delta = 0.80000000000000004 (Default)
kappa = 0.75 (Default)
t0 = 10 (Default)
init_buffer = 75 (Default)
term_buffer = 50 (Default)
window = 25 (Default)
algorithm = hmc (Default)
hmc
engine = nuts (Default)
nuts
max_depth = 10 (Default)
metric = diag_e (Default)
stepsize = 1 (Default)
stepsize_jitter = 0 (Default)
id = 0 (Default)
data
file = BernData.txt
init = 2 (Default)
random
seed = 1724961455
output
file = output.csv (Default)
diagnostic_file = (Default)
refresh = 100 (Default)
Gradient evaluation took 0 seconds
1000 transitions using 10 leapfrog steps per transition would take 0 seconds.
Adjust your expectations accordingly!
Iteration: 1 / 2000 [ 0%] (Warmup)
Iteration: 100 / 2000 [ 5%] (Warmup)
…
Iteration: 1000 / 2000 [ 50%] (Warmup)
Iteration: 1001 / 2000 [ 50%] (Sampling)
Iteration: 1100 / 2000 [ 55%] (Sampling)
…
Iteration: 2000 / 2000 [100%] (Sampling)
# Elapsed Time: 0.016 seconds (Warm-up)
# 0.031 seconds (Sampling)
# 0.047 seconds (Total)
The first block described the options that we chose. We have relied entirely on the defaults. Most of this makes no sense until you understand the sampler that is being used, so we will leave it for the moment. Progress is indicated every 5% of the way through the simulations; warmup is just another term for what everyone else calls the burnin. Finally, we see that the run took under 0.05 seconds once the compilation was complete.
The output file
The file output.csv starts by giving the same information about the options as we find in the log file, then we get the simulated values and we finish with the timings. The central portion has one line for each value of theta but also includes columns giving details of the performance of the samplers.
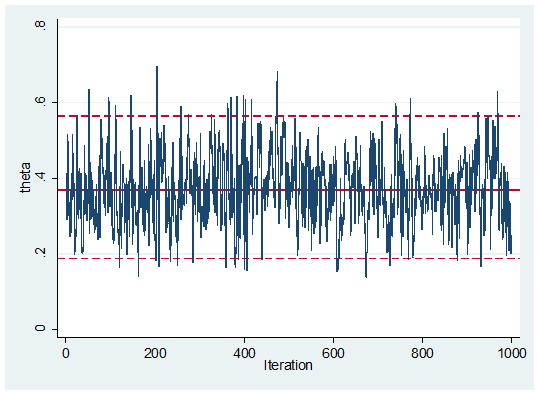
I opened the file in Excel and manually copied and pasted the column of values of theta into Stata. Here is the trace plot
The mixing looks fine although this simple problem is hardly a test of the algorithm.
The summary statistics are,
—————————————————————————-
Parameter n mean sd sem median 95% CrI
—————————————————————————-
theta 1000 0.371 0.099 0.0053 0.369 ( 0.188, 0.565 )
—————————————————————————-
If we had an independent sample, the sem would be 0.099/sqrt(1000)=0.0031 so the autocorrelation has increased the variance by a factor of (0.0053/0.0031)^2=2.9. Not stunningly impressive but, because of the samplers that it uses, we would expect Stan to really shine when we have more complex models with highly correlated parameters. Over the next few weeks we will see if that expectation is justified.


 Subscribe to John's posts
Subscribe to John's posts
Recent Comments