This blog discusses the teaching of data analysis with R. It was inspired by a short course that I first ran in the autumn of 2018. The notes for that course can be found on my github page. If you have not already done so then you might start by reading my introduction to this blog.
In this post I discuss the plan behind the first session of the course.
Since I want to discuss the planning of a session of teaching, it might help to first set out some guiding principles. I think that they are generally applicable, but since I only teach R and biostatistics, I must be careful not to claim too much. Here is a short list
- don’t waste time on facts that the students can find for themselves
- motivate
- explain why we do things as we do
- explain how things work
- emphasise structure
- only teach things that the students need to learn
- simplify, simplify, simplify
The idea that runs through all of these points is that modern teaching should concentrate on those things that you cannot find for yourself on the internet. Things such as, motivation, the reason why we do things as we do and the broader structure within which the specific ideas will apply. After that, avoid digression into things that interest you, but which are not relevant to the students and at every turn, ask yourself whether you could possibly make it any simpler.
Starting with the data forces you into this way of thinking. The data provide the motivation and they help you to keep focus throughout the session. I hope that the impact of my list of principles will be obvious as I discuss the way in which I planned the first session.
This introductory course consisted of four half-day sessions. So, after allowing for breaks, each session had about three hours of contact time. I decided to divide this time so that my lecture and the demonstration of my analysis of the chosen data each had about 45 minutes and the remaining time was left for the students to work on an analysis exercise. As I’ve explained in a previous post, each session is based on a single dataset taken from a published article in the general area of the life sciences. So as always I started by hunting through the dryad repository for a suitable dataset.
Not surprisingly the first session presented me with more problems than any other. I decided to create structure in this session by establishing a full workflow from data import through to report writing and since I could not assume any prior knowledge of R, I needed to find a real study that was simple enough that I could create a meaningful analysis with very basic code.
I found what I thought was a suitable study that investigated the way that lizards adapted to an urban environment by timing them running on different artificial surfaces. Unfortunately, I made the mistake of opting to use the same data for Session 1 and Session 2. My hope had been that this would save time, but in reality it just caused confusion. Session 1 went well, but problems arose in Session 2 when I revisited the analysis from the previous session and made changes to it. Some students confused the code needed for the two analyses. Consequently, the second time that I ran the course, I kept the lizard data for the second session and looked for a replacement dataset for Session 1.
My first attempt to re-write Session 1 was based on a PLoS One paper describing a study in which simple measurements were made on an occupation cohort. I will not identify the article because I discovered something strange about their data. There were lots of duplicates, that is, far too many of the measurements, made on supposedly different subjects, were identical. Even though I had prepared much of the material for my first session based on these data, I decided to abandon it and look for a more suitable study.
My third attempt was based on a study of African Buffalo also published in PLoS One. The study created reference intervals based for biochemical measurements made on blood samples taken from wild buffalo. The idea is that you measure, say, serum calcium, in hundreds of blood samples taken from wild buffalo and then calculate an interval (x, y) such that, in healthy buffalo, 90% of serum calcium measurements are between x and y. In the future, the reference interval can be used to help assess the health of a buffalo. The task is easily understood, the analysis that can be completed using simple descriptive statistics and I was able to demonstrate the analysis with one biochemical measurement and then invite the students to looks at another in the exercise.
The complication with this particular study was that the researchers captured wild buffalo and kept them in a large enclosure, so that they remained free but could be recaptured every three months in order to have blood samples taken. As a result, there were about 450 blood samples from around 100 animals. The article did not adjust its analysis for this complication and when I re-analysed the data, I found that an adjustment would have made very little difference.
An appreciation of the implications of having repeated measurements on the same individual marks a big step in someone’s understanding of statistics, but it is not a topic that I wanted to become involved in during my very first session. After some hesitation, I decided to follow the article and use all of the measurements without adjustment, but I added a question in the exercise that hints at the complication. That way, if any of the students had enough background knowledge to appreciate the consequences of re-sampling the same animals, I could spend time discussing statistical independence with them without involving the students who were operating at a more basic level.
Session 1 is particularly challenging because of the breadth of material that needs to be included in order to cover the workflow of a complete data analysis. As I always do, I started my preparation by analysing the data for myself, so as to identify the skills that I would need to teach in that lecture.
In the case of the African buffalo, the data are available on dryad in a csv file, so I knew that I needed to teach read_csv(), which is the tidyverse’s update of base R’s read.csv() function. By default, read_csv() reads the data as either numeric or character, so I did not need to explain the use of factors or the horrid stringsAsFactors argument. When I analyse data, I always import it and then immediately save it in rds format. I think that this is good practice, so I wanted to start the students working in that way, which meant covering write_rds() and read_rds() in the lecture.
Unfortunately, the dataset includes a single biochemical measurement of “<2”. I never doctor the data for my teaching and the principle of reproducibility, excludes the possibility of editing the data interactively, so I knew that I would need to find a way of handling the awkward observation using very simple R code.
Data editing in the tidyverse is not as neat as perhaps it ought to be given how basic the operation is. Data checking and data cleaning are important aspects of any analysis, but they have been rather neglected by the tidyverse. Editing involves changes to selected entries in a column, the sort of thing that one would do with subscripts in base R. You can, of course, combine mutate() with ifelse() or replace(), but I did not want to get into that level of coding in the first session. Rather confusingly, I think, mutate_if() will edit selected columns, but will not perform the more common task of editing selected rows, perhaps a mutate_where() function would be a useful addition to dplyr, or better still a ‘where’ argument could be added to mutate(). In the end, I opted for the simple solution of reading the column containing “<2” as a character variable and using the as.numeric() function within mutate() to convert to a numeric variable. This function replaces “<2” by a missing value.
I did not want to talk about analysing missing data in the first session, so I avoided it by choosing not to analyse the biochemical with the “<2” entry. Since I was not planning to use that variable, I could have left it as a character variable, but one of the things that I like to stress is the importance of checking what you do. I bang on repeatedly about how data analysis is complex, so errors are are inevitable; consequently the important thing is not the error, but whether you spot it. Importing data provides a good opportunity to talk about this basic principle and one cannot notice a problem and then do nothing about it.
The csv file on dryad was probably exported from Excel because it has some column names that include spaces, such as “Animal ID”. So there was no way around teaching the use of single sloping quotes around column names, as in `Animal ID`. Not something that you would usually associate with Session 1 of an R course. I use lowerCaseCamel for my variable names, so to be able to turn `Animal ID` into animalId, I needed to teach the rename() function as well as the use of quoted variable names. I decided to cover rename() in the lecture, but I did not mention the use of quotes around variable names until they were needed in the demonstration. Later in the course, I introduce the janitor package and its excellent clean_names() function.
With this style of teaching, one does not need to cover every point in the lecture. The lecture is for establishing the basic syntax, but coding methods needed for the specific dataset can still be left to the demonstration. It is not necessary to give a comprehensive account of each function that you use, you just give the detail needed for the problem at hand. The medium term aim is to get the student to the point where the lecturer gives them the basic syntax and they look up the particulars needed to solve a specific coding problem. With this in mind, it is a good idea to include questions in the exercise that require the student to discover for themselves, how to code variants of the functions that they have been taught. At first, one can help by suggesting a webpage with relevant examples, but eventually the students need to be self-sufficient and to search for a solution for themselves. A couple of the tasks in the exercise for Session 1 include links to helpful webpages, although at this stage, they provide extra information that is not essential to completing the exercise.
For my analysis of the buffalo data I needed to create summary statistics using summarise() from dplyr and the base R function quantile(). When I analysed the data, I needed to check whether a reference interval depends on factors such as the gender of the buffalo, or the season of the year when the blood sample was taken, which I could do with group_by(). Gender is saved by read_csv() as a character variable rather than a factor, but this does not matter to group_by() as it works equally well with character strings. I also needed the function filter() to pick out the first measurement on each animal as a way of investigating the impact of repeat measurements.
For visualisation, I used histograms of the different biochemical measurements and since the dataset also contains the age of the animal at the time of each measurement, I was able to employ a scatter plot of measurement against age. Both types of plot can be created with qplot() without the need to know anything of the ggplot syntax. Here I made a point of being honest with the students by telling them that qplot() would be dropped after the first session in favour of the much more comprehensive capabilities of ggplot2.
Finally, I wanted to create a report based on the analysis using rmarkdown. This first report was restricted to text, section headers and R chunks and was knitted into an html document. This is far easier to demonstrate than it is to lecture, so I left this until the demonstration in which I took RStudio’s dummy document and striped out everything except the YAML header before adding my own content. I only needed chunk options to avoid the unsightly information that R provides when the tidyverse is loaded; perhaps the authors of rmarkdown should consider changing the defaults for chunk options warning and message to FALSE.
When the lecture is planned around what is actually needed for a particular analysis, the amount of factual information that needs to be taught is often surprisingly small. This frees you to concentrate on the principles of R coding, effectively an explanation of how R works. So, on top of the functions needed for the analysis, I also able to talk about,
- the pipe, %>%
- the installation and loading of packages
- the idea of a data frame
- the benefits of saving data in rds format
I could happily spend a hour on each if these topics, but in my lecture I cover each one in under five minutes. For instance, there are good reasons for preferring rds format to the alternative rdata format, but most of my students will never have used rdata, so the comparison is irrelevant to them. The important learning point for the first session is, why it is a good idea to save the data in binary format. Many of the students are used to starting each day by re-reading and re-tidying the raw data, a process that is wasteful and which invites errors and inconsistencies.
In the demonstration, I created an analysis of the serum calcium measurements. I tried to do this in a structured way by having stages in which the data are imported and tidied, analysed and then reported on. To encourage the students to think about the workflow behind an analysis, I created a diagram using the R package diagrammeR.
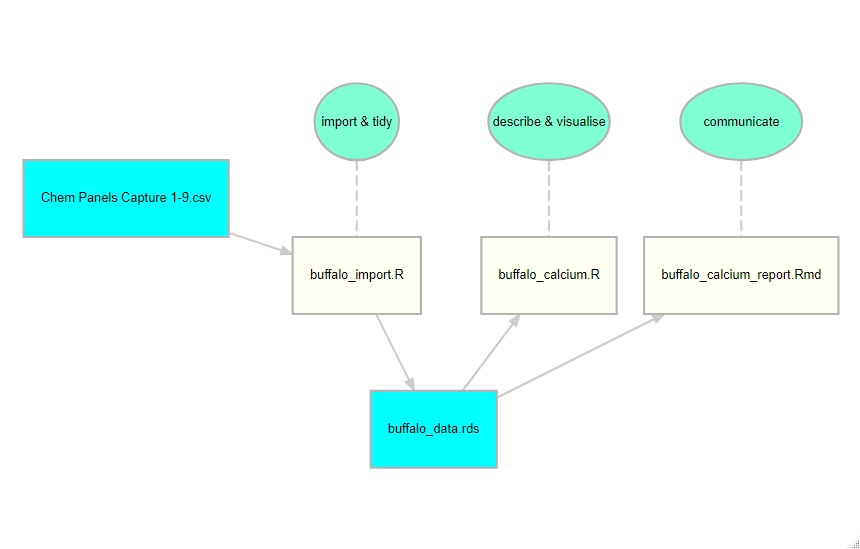
The diagram shows that the data are read from a csv file called ‘Chem Panels Captures 1-9.csv’ and tidied with the script, buffalo_import.R. The resulting data are saved as buffalo_data.rds. My analysis of the blood calcium measurements is coded in the script, buffalo_calcium.R, and I write a report summarising my analysis with buffalo_calcium_report.rmd.
When I first ran the course, I did not provide this diagram and I was genuinely surprised that students lost track of what we were doing at each stage. This was clearly my problem and not theirs. I am used to large analyses that have to be divided into stages, while they are not; indeed, many of them are new to working with scripts rather than interactive commands or point and click. Foolishly, I made things even worse by having separate scripts of import and tidying. This reflects my own way of working; routinely, I start by importing everything and saving it immediately in rds format, then I tidy it and save the tidied version. This is quite sensible when you have large complex datasets but totally unnecessary when you are just starting out, so second time around I combined the import and tidying into a single script.
One advantage of splitting the analysis across scripts was that in the exercise the students were asked to create reference intervals for serum phosphorus and they were able to use buffalo_import.R virtually unedited. The key idea that I was trying to get across was that once you have a structure for your work, you can easily slot new components into it.
The handout that I provide for the students can be downloaded from my github pages. It contains notes on the material covered in the lecture, the code that I use in the demonstration and the question in the exercise. I also provide my students with the demonstration R scripts as separate files so that those who wish, can mirror my demonstration in real time on their own computer. These notes are intended to be a reference for the students and although they correspond quite closely to my lecture, they are not exactly equivalent. For instance, I mention rmarkdown in the notes, but I do not talk about it in the lecture, but instead I leave it to the demonstration.
If you look at my R scripts for the demonstration you will notice some points of personal style that are not standard practice. These include
- my style for adding comments to the scripts
- my preference for adding DF at the ends of the names of data frames
- my liking for the right pointing arrow, ->
- my habit of placing a block of code near the start of the script in which I define the project folder (home) and all filenames
I stress in the lecture that these are personal choices that the students do not need to follow. All I insist on is literacy and consistency of whatever style they choose. In practice, almost all students copy my style, at least for the first couple of sessions.
The exercise is the third part of the session and it asks the students to produce a similar analysis of the serum phosphorus measurements. I timetable it so that they have about one and a half hours to work on the exercise. Not all complete it in that time, but most get close.
I provide quite comprehensive notes on the solution to the exercise. With other courses, I have tried making the solution available at the end of the session, but I’ve come to the conclusion that it is better to release it prior to the course, together with all of the other course material. The danger of early release is that weak students attempt the exercise by copying and pasting code from my solution. This is doubly frustrating, not only do they learn very little, but they walk away with the idea that there is a correct solution that they were meant to arrive at. However, keeping back the solution stops students from checking their answers and sometimes they waste time over minor points of coding. Struggling with coding is an important part of learning R, but it is not so important that I want a student to take a hour of valuable class time over something trivial; of course, they should ask for help, but some people find that difficult, especially when they do not know the lecturer and unfortunately, we are not so well resourced with tutors that we can always offer immediate assistance. Providing the solution in advance frees up my time, so that instead of explaining the same piece of coding over and over again, I am free to go around and discuss the broader objectives of the analysis at a level appropriate to each student.
Feedback from the students was generally positive. The students on the course were all junior researchers who needed to analyse their own data and who had chosen to come on the course; they were not being assessed. Of course, there were a few who decided that R is not for them, but most students got a lot from this session. One student who had previously suffered on a more traditional R course, even told me that he could not believe that R was that easy.

 Subscribe to John's posts
Subscribe to John's posts
Recent Comments